The n_components parameter in scikit-learn’s LinearDiscriminantAnalysis controls the number of components for dimensionality reduction.
Linear Discriminant Analysis (LDA) is a method used for classification and dimensionality reduction. It projects the data onto a lower-dimensional space while maximizing the separability between classes.
The n_components parameter determines the number of components to keep after the LDA transformation. It affects the model’s ability to capture class distinctions and can impact classification performance.
By default, n_components is set to min(n_classes - 1, n_features), where n_classes is the number of target classes and n_features is the number of input features.
In practice, values ranging from 1 to min(n_classes - 1, n_features) are commonly used, depending on the dataset’s characteristics and the desired trade-off between dimensionality reduction and class separability.
from sklearn.datasets import make_classification
from sklearn.model_selection import train_test_split
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
from sklearn.metrics import accuracy_score
import numpy as np
import matplotlib.pyplot as plt
# Generate synthetic dataset
X, y = make_classification(n_samples=1000, n_features=20, n_informative=15,
n_redundant=5, n_classes=5, random_state=42)
# Split into train and test sets
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
# Train with different n_components values
n_components_range = range(1, 5) # 5 classes, so max n_components is 4
accuracies = []
for n in n_components_range:
lda = LinearDiscriminantAnalysis(n_components=n)
lda.fit(X_train, y_train)
X_train_lda = lda.transform(X_train)
X_test_lda = lda.transform(X_test)
# Train a new LDA classifier on the transformed data
lda_clf = LinearDiscriminantAnalysis()
lda_clf.fit(X_train_lda, y_train)
y_pred = lda_clf.predict(X_test_lda)
accuracy = accuracy_score(y_test, y_pred)
accuracies.append(accuracy)
print(f"n_components={n}, Accuracy: {accuracy:.3f}")
# Plot accuracy vs n_components
plt.plot(n_components_range, accuracies, marker='o')
plt.xlabel('n_components')
plt.ylabel('Accuracy')
plt.title('LDA Performance vs n_components')
plt.show()
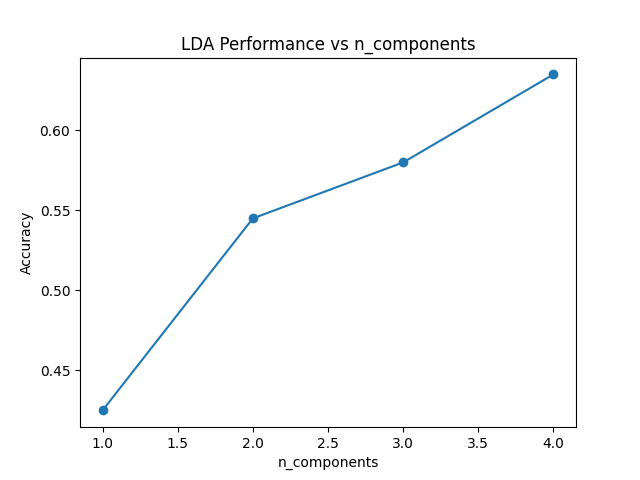
Running the example gives an output like:
n_components=1, Accuracy: 0.425
n_components=2, Accuracy: 0.545
n_components=3, Accuracy: 0.580
n_components=4, Accuracy: 0.635
The key steps in this example are:
- Generate a synthetic multi-class dataset with informative and redundant features
- Split the data into train and test sets
- Train
LinearDiscriminantAnalysismodels with differentn_componentsvalues - Transform the data using LDA and train a classifier on the transformed data
- Evaluate the accuracy of each model on the test set
- Visualize the relationship between
n_componentsand classification accuracy
Tips for setting n_components:
- Start with the default value and experiment with lower values to find the optimal trade-off between dimensionality reduction and classification performance
- Consider the number of classes in your dataset, as
n_componentscannot exceed n_classes - 1 - Examine the explained variance ratio to determine how much information is retained by each component
Issues to consider:
- Using too few components may result in loss of important class-discriminating information
- Using too many components may lead to overfitting, especially with small datasets
- The optimal
n_componentsvalue depends on the specific characteristics of your dataset and the problem you’re trying to solve
